3 Multi-Row Headers
- Lisa DeBruine (2021-10-17)
A student on our help forum once asked for help making a wide-format dataset long. When I tried to load the data, I realised the first three rows were all header rows. Here's the code I wrote to deal with it.
First, I'll make a small CSV "file" below. In a typical case, you'd read the data in from a file.
demo_csv <- I("SUB1, SUB1, SUB1, SUB1, SUB2, SUB2, SUB2, SUB2
COND1, COND1, COND2, COND2, COND1, COND1, COND2, COND2
X, Y, X, Y, X, Y, X, Y
10, 15, 6, 2, 42, 4, 32, 5
4, 43, 7, 34, 56, 43, 2, 33
77, 12, 14, 75, 36, 85, 3, 2")If you try to read in this data, you'll get a message about the duplicate column names and the resulting table will have "fixed" column headers and the next two columns headers as the first two rows.
data <- read_csv(demo_csv)| SUB1...1 | SUB1...2 | SUB1...3 | SUB1...4 | SUB2...5 | SUB2...6 | SUB2...7 | SUB2...8 |
|---|---|---|---|---|---|---|---|
| COND1 | COND1 | COND2 | COND2 | COND1 | COND1 | COND2 | COND2 |
| X | Y | X | Y | X | Y | X | Y |
| 10 | 15 | 6 | 2 | 42 | 4 | 32 | 5 |
| 4 | 43 | 7 | 34 | 56 | 43 | 2 | 33 |
| 77 | 12 | 14 | 75 | 36 | 85 | 3 | 2 |
Instead, you should read in just the header rows by setting n_max equal to the number of header rows and col_names to FALSE.
data_head <- read_csv(demo_csv,
n_max = 3,
col_names = FALSE)You will get a table that looks like this:
| X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 |
|---|---|---|---|---|---|---|---|
| SUB1 | SUB1 | SUB1 | SUB1 | SUB2 | SUB2 | SUB2 | SUB2 |
| COND1 | COND1 | COND2 | COND2 | COND1 | COND1 | COND2 | COND2 |
| X | Y | X | Y | X | Y | X | Y |
You can then make new header names by pasting together the names in the three rows by summarising across all the columns with the paste() function and collapsing them using "_". Use unlist() and unname() to convert the result from a table to a vector.
new_names <- data_head %>%
summarise(across(.fns = paste, collapse = "_")) %>%
unlist() %>% unname()
new_names## [1] "SUB1_COND1_X" "SUB1_COND1_Y" "SUB1_COND2_X" "SUB1_COND2_Y" "SUB2_COND1_X"
## [6] "SUB2_COND1_Y" "SUB2_COND2_X" "SUB2_COND2_Y"Now you can read in the data without the three header rows. Use skip to skip the headers and set col_names to the new names.
data <- read_csv(demo_csv,
skip = 3,
col_names = new_names,
show_col_types = FALSE)| SUB1_COND1_X | SUB1_COND1_Y | SUB1_COND2_X | SUB1_COND2_Y | SUB2_COND1_X | SUB2_COND1_Y | SUB2_COND2_X | SUB2_COND2_Y |
|---|---|---|---|---|---|---|---|
| 10 | 15 | 6 | 2 | 42 | 4 | 32 | 5 |
| 4 | 43 | 7 | 34 | 56 | 43 | 2 | 33 |
| 77 | 12 | 14 | 75 | 36 | 85 | 3 | 2 |
If you have an excel file that merges the duplicate headers across rows, it's a little trickier, but still do-able.
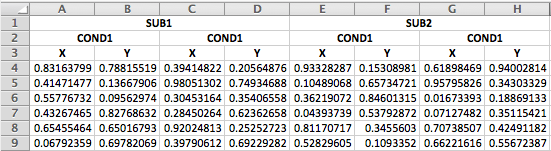
The first steps is the same: read in the first three rows.
data_head <- read_excel("data/3headers_demo.xlsx",
n_max = 3,
col_names = FALSE)| ...1 | ...2 | ...3 | ...4 | ...5 | ...6 | ...7 | ...8 |
|---|---|---|---|---|---|---|---|
| SUB1 | NA | NA | NA | SUB2 | NA | NA | NA |
| COND1 | NA | COND2 | NA | COND1 | NA | COND2 | NA |
| X | Y | X | Y | X | Y | X | Y |
The code below starts at the second column and fills in any missing data with the value in the previous column.
for (i in 2:ncol(data_head)) {
prev <- data_head[, i-1]
this <- data_head[, i]
missing <- is.na(this)
this[missing, ] <- prev[missing, ]
data_head[, i] <- this
}Now you can continue generating the pasted name the same as above.
new_names <- data_head %>%
summarise(across(.fns = paste, collapse = "_")) %>%
unlist() %>% unname()
new_names## [1] "SUB1_COND1_X" "SUB1_COND1_Y" "SUB1_COND2_X" "SUB1_COND2_Y" "SUB2_COND1_X"
## [6] "SUB2_COND1_Y" "SUB2_COND2_X" "SUB2_COND2_Y"If your data are set up with multiple headers, you'll probably want to change the shape of the data. Here's a quick example how to use pivot_longer() and pivot_wider() to do this with variable names like above.
data <- read_excel("data/3headers_demo.xlsx",
skip = 3,
col_names = new_names)
data_long <- data %>%
# add a row ID column if one doesn't exist already
mutate(trial_id = row_number()) %>%
# make a row for each data column
pivot_longer(
cols = -trial_id, # everything except trial_id
names_to = c("sub_id", "condition", "coord"),
names_sep = "_",
values_to = "val"
) %>%
# make x and y coord columns
pivot_wider(
names_from = coord,
values_from = val
)| trial_id | sub_id | condition | X | Y |
|---|---|---|---|---|
| 1 | SUB1 | COND1 | 0.8316380 | 0.7881552 |
| 1 | SUB1 | COND2 | 0.3941482 | 0.2056488 |
| 1 | SUB2 | COND1 | 0.9332829 | 0.1530898 |
| 1 | SUB2 | COND2 | 0.6189847 | 0.9400281 |
| 2 | SUB1 | COND1 | 0.4147148 | 0.1366791 |
| 2 | SUB1 | COND2 | 0.9805130 | 0.7493469 |
| 2 | SUB2 | COND1 | 0.1048907 | 0.6573472 |
| 2 | SUB2 | COND2 | 0.9579583 | 0.3430333 |
| 3 | SUB1 | COND1 | 0.5577673 | 0.0956297 |
| 3 | SUB1 | COND2 | 0.3045316 | 0.3540656 |
| 3 | SUB2 | COND1 | 0.3621907 | 0.8460132 |
| 3 | SUB2 | COND2 | 0.0167339 | 0.1886913 |
| 4 | SUB1 | COND1 | 0.4326746 | 0.8276863 |
| 4 | SUB1 | COND2 | 0.2845026 | 0.6236266 |
| 4 | SUB2 | COND1 | 0.0439374 | 0.5379287 |
| 4 | SUB2 | COND2 | 0.0712748 | 0.3511542 |
| 5 | SUB1 | COND1 | 0.6545546 | 0.6501679 |
| 5 | SUB1 | COND2 | 0.9202481 | 0.2525272 |
| 5 | SUB2 | COND1 | 0.8117072 | 0.3455603 |
| 5 | SUB2 | COND2 | 0.7073851 | 0.4249118 |
| 6 | SUB1 | COND1 | 0.0679236 | 0.6978207 |
| 6 | SUB1 | COND2 | 0.3979061 | 0.6922928 |
| 6 | SUB2 | COND1 | 0.5282960 | 0.1093352 |
| 6 | SUB2 | COND2 | 0.6622162 | 0.5567239 |